Fighting cancer: simulations at unprecedented length- and time-scales
See Machine learning–driven multiscale modeling reveals lipid-dependent dynamics of RAS signaling proteins. More info.
Biochemical and Biophysical Systems Group |


The Biochemical and Biophysical Systems Group’s scientists use cutting-edge, multi-scale, in silico simulations to tackle problems in biology. We use a wide range of computational biology methods that employ LLNL’s high-performance computing resources to simulate systems from sub-atomic scale to population level. These methods include atomistic and coarse-grained molecular dynamics, quantum simulations, constraint-based genome-scale simulations, reaction-transport dynamic simulations, and agent-based, whole-organ, and pharmacokinetics/pharmacodynamics models. We develop new computational methods to describe and predict biological systems. In addition, we combine experimental efforts with physics-based simulations and statistical and machine-learning models to accelerate the design and development of safe and effective therapeutics. Overall, we seek predictive understanding of protein-mediated processes related to critical missions of LLNL, including bioenergy, medical countermeasures, and new materials.
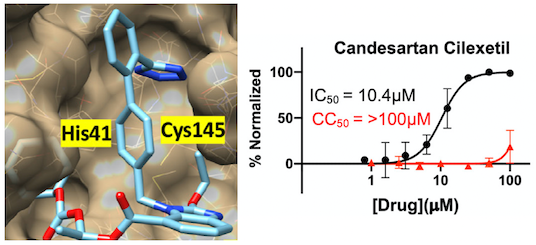
Discovery of BBO-8520, a first-in-class direct and covalent dual inhibitor of GTP-bound (ON) and GDP-bound (OFF) KRASG12C
Anna E. Maciag, James P. Stice, Bin Wang, Alok K. Sharma, Albert H. Chan, Ken Lin, Devansh Singh, Marcin Dyba, Yue Yang, Saman Setoodeh, Brian P. Smith, Jin Hyun Ju, Stevan Jeknic, Dana Rabara, Zuhui Zhang, Erik K. Larsen, Dominic Esposito, John-Paul Denson, Michela Ranieri, Mary Meynardie, Sadaf Mehdizadeh, Patrick A. Alexander, Maria Abreu Blanco, David M. Turner, Rui Xu, Felice C. Lightstone, Kwok-Kin Wong, Andrew G. Stephen, Keshi Wang, Dhirendra K. Simanshu, Kerstin W. Sinkevicius, Dwight V. Nissley, Eli Wallace, Frank McCormick, Pedro J. Beltran
Cancer Discovery (2024)
Targeting Ras-, Rho-, and Rab-family GTPases via a conserved cryptic pocket
Johannes Morstein, Victoria Bowcut, Micah Fernando, Yue Yang, Lawrence Zhu, Meredith L. Jenkins, John T. Evans, Keelan Z. Guiley, D. Matthew Peacock, Sophie Krahnke, Zhi Lin, Katrine A. Taran, Benjamin J. Huang, Andrew G. Stephen, John E. Burke, Felice C. Lightstone, Kevan M. Shokat
Cell (2024)
Transporter annotations are holding up progress in metabolic modeling
John Casey, Brian Bennion, Patrik D’haeseleer, Jeffrey Kimbrel, Gianna Marschmann and Ali Navid
Frontiers in Systems Biology 4 (2024)
Building complex membranes with Martini 3
Tugba Nur Ozturk, Melanie König, Timothy S. Carpenter, Kasper B. Pedersen, Tsjerk A. Wassenaar, Helgi I. Ingólfsson, Siewert J. Marrink
Methods in Enzymology (2024)
Spatially structured competition and cooperation alters algal carbon flow to bacteria
Hyungseok Kim, Vanessa L. Brisson, John R. Casey, Courtney Swink, Kristina A. Rolison, Amber N. Golini, Trent R. Northen, Peter K. Weber, Dušan Veličković, Cullen R. Buie, Xavier Mayali, Rhona K. Stuart
bioRxiv (2024)
Disentangling top-down drivers of mortality underlying diel population dynamics of Prochlorococcus in the North Pacific Subtropical Gyre
Stephen J. Beckett, David Demory, Ashley R. Coenen, John R. Casey, Mathilde Dugenne, Christopher L. Follett, Paige Connell, Michael C. G. Carlson, Sarah K. Hu, Samuel T. Wilson, Daniel Muratore, Rogelio A. Rodriguez-Gonzalez, Shengyun Peng, Kevin W. Becker, Daniel R. Mende, E. Virginia Armbrust, David A. Caron, Debbie Lindell, Angelicque E. White, François Ribalet & Joshua S. Weitz
Nature Communications (2024) 15, 2105.
Enhancing Docking Accuracy with PECAN2, a 3D Atomic Neural Network Trained without Co-Complex Crystal Structures
Heesung Shim, Jonathan E. Allen, W.F. Drew Bennett
Machine Learning and Knowledge Extraction (2024) 6(1), 642-657.
CyanoCyc cyanobacterial web portal
Lisa R. Moore, Ron Caspi, Douglas A. Campbell, John R. Casey, Sophie Crevecoeur, David J. Lea-Smith, Bin Long, Naaman M. Omar, Suzanne M. Paley, Nicolas M. Schmelling, Alejandro Torrado, Jonathan P. Zehr, and Peter D. Karp
Frontiers in Microbiology (2024)15:1340413.
Advances in Computational Approaches for Estimating Passive Permeability in Drug Discovery
Austen Bernardi, W. F. Drew Bennett, Stewart He, Derek Jones, Dan Kirshner, Brian J. Bennion and Timothy S. Carpenter
Membranes (2023) 13 851.
Clustering Protein Binding Pockets and Identifying Potential Drug Interactions: A Novel Ligand-Based Featurization Method
Garrett A. Stevenson, Dan Kirshner, Brian J. Bennion, Yue Yang, Xiaohua Zhang, Adam Zemla, Marisa W. Torres, Aidan Epstein, Derek Jones, Hyojin Kim, W. F. Drew Bennett, Sergio E. Wong, Jonathan E. Allen, Felice C. Lightstone
Journal of Chemical Information and Modeling (2023) 63, 21, 6655-6666.
Multiscale simulations reveal TDP-43 molecular level interactions driving condensation
Helgi I. Ingólfsson, Azamat Rizuan, Xikun Liu, Priyesh Mohanty, Paulo C.T. Souza, Siewert J. Marrink, Michael T. Bowers, Jeetain Mittal, Joel Berry
Biophysical Journal (2023).
Evaluating point-prediction uncertainties in neural networks for protein-ligand binding prediction
Ya Ju Fan, Jonathan E. Allen, Kevin S. McLoughlin, Da Shi, Brian J. Bennion, Xiaohua Zhang, Felice C. Lightstone
Artificial Intelligence Chemistry (2023) 1 (1).
Machine Learning-driven Multiscale Modeling: Bridging the Scales With a Next Generation Simulation Infrastructure
Helgi I. Ingólfsson, Harsh Bhatia, Fikret Aydin, Tomas Oppelstrup, Cesar A López, Liam G. Stanton, Timothy S. Carpenter, Sergio Wong, Francesco Di Natale, Xiaohua Zhang, Joseph Y. Moon, Christopher B. Stanley, Joseph R. Chavez, Kien Nguyen, Gautham Dharuman, Violetta Burns, Rebika Shrestha, Debanjan Goswami, Gulcin Gulten, Que N. Van, Arvind Ramanathan, Brian Van Essen, Nicolas W. Hengartner, Andrew G. Stephen, Thomas Turbyville, Peer-Timo Bremer, S. Gnanakaran, James N. Glosli, Felice C. Lightstone, Dwight V. Nissley, Frederick H. Streitz
J. Chem. Theory Comput. (2023) 19 (9), 2658-2675.
The confluence of machine learning and multiscale simulations
Harsh Bhatia, Fikret Aydin, Timothy S. Carpenter, Felice C. Lightstone, Peer-Timo Bremer, Helgi I. Ingólfsson, Dwight V. Nissley, Frederick H. Streitz
Current Opinion in Structural Biology (2023) 80, 102569.
Dynamic density functional theory of multicomponent cellular membranes
Liam G. Stanton, Tomas Oppelstrup, Tim S. Carpenter, Helgi I. Ingólfsson, Michael P. Surh, Felice C. Lightstone, and James N. Glosli
Physical Review Research (2023) 5, 013080.
Electrogenetic signaling and information propagation for controlling microbial consortia via programmed lysis
Eric VanArsdale, Ali Navid, Monica J. Chu, Tiffany M. Halvorsen, Gregory F. Payne, Yongqin Jiao, William E. Bentley, Mimi C. Yung
Biotechnology and Bioengineering (2023) 1-16.
PDBspheres – a method for finding 3D similarities in local regions in proteins
Adam Zemla, Jonathan E Allen, Dan Kirshner, Felice C Lightstone
NAR Genomics and Bioinformatics (2022) 4 (4), lqac078.
Scalable Composition and Analysis Techniques for Massive Scientific Workflows
Dong H. Ahn, Xiaohua Zhang, Jeffrey Mast, Stephen Herbein, Francesco Di Natale, Dan Kirshner, Sam Ade Jacobs, Ian Karlin, Daniel J. Milroy, Bronis De Supinski, Brian Van Essen, Jonathan Allen, Felice C. Lightstone
IEEE 18th International Conference on e-Science (2022) 32-43.
(Won Best Paper)
Deep Generative Molecular Design Reshapes Drug Discovery
Xiangxiang Zeng, Fei Wang, Yuan Luo, Seung-gu Kang, Jian Tang, Felice C. Lightstone, Evandro F. Fang, Wendy Cornell, Ruth Nussinov, Feixiong Cheng
Cell Reports Medicine (2022) 3, 100794.
Exploring CRD mobility during RAS/RAF engagement at the membrane
Kien Nguyen, Cesar A. López, Chris Neale, Que N. Van, Timothy S. Carpenter, Francesco Di Natale, Timothy Travers, Timothy H. Tran, Albert H. Chan, Harsh Bhatia, Peter H. Frank, Marco Tonelli, Xiaohua Zhang, Gulcin Gulten, Tyler Reddy, Violetta Burns, Tomas Oppelstrup, Nick Hengartner, Dhirendra K Simanshu, Peer-Timo Bremer, De Chen, James N. Glosli, Rebika Shrestha, Thomas Turbyville, Frederick H. Streitz, Dwight V. Nissley, Helgi I. Ingólfsson, Andrew G. Stephen, Felice C. Lightstone, Sandrasegaram Gnanakaran
Biophysical Journal (2022) 121 (19), 3630.
A Biology-Informed Similarity Metric for Simulated Patches of Human Cell Membrane
Harsh Bhatia, Jayaraman J. Thiagarajan, Rushil Anirudh, T. S. Jayram, Tomas Oppelstrup, Helgi I. Ingólfsson, Felice C. Lightstone, Peer-Timo Bremer
Machine Learning: Science and Technology (2022) 3 (3), 2632-2153.
Bacterial exometabolites influence Chlamydomonas cell cycle and double algal productivity
Miriam Windler, Rhona Stuart, Joerg S. Deutzmann, Xavier Mayali, Ali Navid, Patrik D’haeseleer, Oana E. Marcu, Mary Lipton, Carrie Nicora, Alfred M. Spormann
FEMS Microbiology Ecology (2022) 98, 1.
On the Rapid Calculation of Binding Affinities for Antigen and Antibody Design and Affinity Maturation Simulations
Simone Conti, Edmond Y. Lau, and Victor Ovchinnikov (2022)
Antibodies (2022) 11(3), 51.
Asynchronous Reciprocal Coupling of Martini 2.2 Coarse-Grained and CHARMM36 All-Atom Simulations in an Automated Multiscale Framework
Cesar A. Lopez, Xiaohua Zhang, Fikret Aydin, Rebika Shrestha, Que N. Van, Christopher B. Stanley, Timothy S. Carpenter, Kien Nguyen, Lara A. Patel, De Chen, Violetta Burns, Nicolas W. Hengartner, Tyler J. E. Reddy, Harsh Bhatia, Francesco Di Natale, Timothy H. Tran, Albert H. Chan, Dhirendra K. Simanshu, Dwight V. Nissley, Frederick H. Streitz, Andrew G. Stephen, Thomas J. Turbyville, Felice C. Lightstone, Sandrasegaram Gnanakaran, Helgi I. Ingolfsson, Chris Neale
Journal of Chemical Theory and Computation (2022) 18 (8), 5025-5045.
Ras-mutant cancers are sensitive to small molecule inhibition of V-type ATPases in mice
Tolani, B., Celli, A., Yao, Y. , Tan, Y.Z., Fetter, R., de Smith, A.J., Vasanthakumar, T., Bisignano, P. et al.
Nat Biotechnol (2022).
Large-scale application of free energy perturbation calculations for antibody design
Fangqiang Zhu, Feliza A. Bourguet, W. F. Drew Bennett, Edmond Y. Lau, Kathryn T. Arrildt, Brent W. Segelke, Adam T. Zemla, Thomas A. Desautels, and Daniel M. Faissol
Scientific Reports (2022) 12, 12489.
Mitochondrial uncouplers induce proton leak by activating AAC and UCP1
Bertholet, A.M., Natale, A.M., Bisignano, P. et al.
Nature 606, 180–187 (2022).
Bacterial Membranes Are More Perturbed by the Asymmetric Versus Symmetric Loading of Amphiphilic Molecules
W. F. Drew Bennett, Stephen J. Fox, Delin Sun, and C. Mark Maupin
Membranes (2022) 12, 4, 350
Regulation of Gramicidin Channel Function Solely by Changes in Lipid Intrinsic Curvature
Andreia M. Maer, Radda Rusinova, Lyndon L. Providence, Helgi I. Ingólfsson, Shemille A. Collingwood, Jens A. Lundbæk and Olaf S. Andersen
Frontiers in Physiology (2022) 13:836789.
Interpretable artificial intelligence and exascale molecular dynamics simulations to reveal kinetics: Applications to Alzheimer's disease
William Martin, Gloria Sheynkman, Felice C Lightstone, Ruth Nussinov, Feixiong Cheng
Current Opinion in Structural Biology (2022) 72, 103.
Multiframe Imaging of Micron and Nanoscale Bubble Dynamics
Garth C. Egan, Edmond Y. Lau, and Eric Schwegler
Nano Letters (2022) 22 (3), 1053-1058
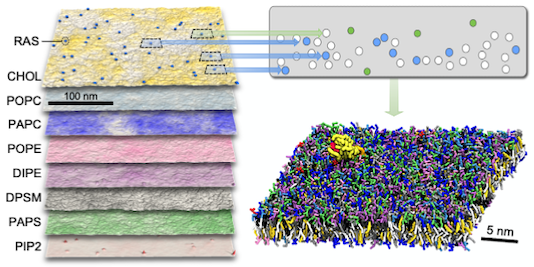
Machine learning–driven multiscale modeling reveals lipid-dependent dynamics of RAS signaling proteins
Helgi I. Ingólfsson, Chris Neale, Timothy S. Carpenter, Rebika Shrestha, Cesar A. López, Timothy H. Tran, Tomas Oppelstrup, Harsh Bhatia, Liam G. Stanton, Xiaohua Zhang, Shiv Sundram, Francesco Di Natale, Animesh Agarwal, Gautham Dharuman, Sara I. L. Kokkila Schumacher, Thomas Turbyville, Gulcin Gulten, Que N. Van, Debanjan Goswami, Frantz Jean-Francois, Constance Agamasu, De Chen, Jeevapani J. Hettige, Timothy Travers, Sumantra Sarkar, Michael P. Surh, Yue Yang, Adam Moody, Shusen Liu, Brian C. Van Essen, Arthur F. Voter, Arvind Ramanathan, Nicolas W. Hengartner, Dhirendra K. Simanshu, Andrew G. Stephen, Peer-Timo Bremer, S. Gnanakaran, James N. Glosli, Felice C. Lightstone, Frank McCormick, Dwight V. Nissley, and Frederick H. Streitz
Proceedings of the National Academy of Sciences (2022) 119 (1) e2113297119.

Copyright © 2024, Lawrence Livermore National Laboratory

